d a t a / s o f t w a r e
data
300 ROI Set
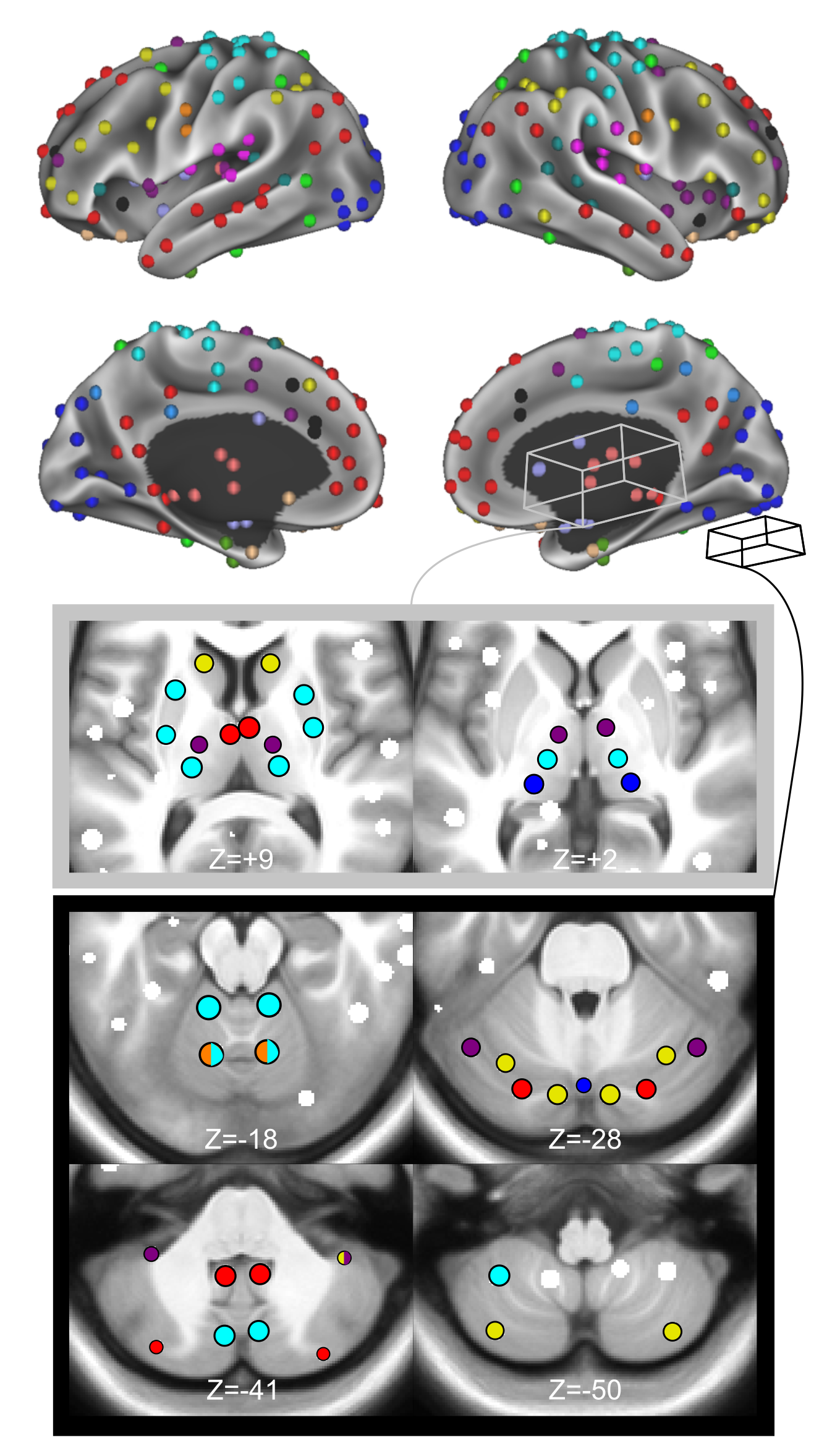
An important aspect of network-based analysis is robust node definition. This issue is critical for functional brain network analyses, as poor node choice can lead to spurious findings and misleading inferences about functional brain organization. Two sets of functional brain nodes from our group are well represented in the literature: (1) 264 volumetric regions of interest (ROIs) reported in Power et al., 2011 and (2) 333 surface parcels reported in Gordon et al., 2016. However, subcortical and cerebellar structures are either incompletely captured or missing from these ROI sets. Therefore, properties of functional network organization involving the subcortex and cerebellum may be underappreciated thus far. Here, we apply a winner-take-all partitioning method to resting-state fMRI data and careful consideration of anatomy to generate novel functionally-constrained ROIs in the thalamus, basal ganglia, amygdala, hippocampus, and cerebellum. We validate these ROIs in three datasets via several anatomical and functional criteria, including known anatomical divisions and functions, as well as agreement with existing literature. Further, we demonstrate that combining these ROIs with established cortical ROIs recapitulates and extends previously described functional network organization. This new set of ROIs is made publicly available for general use, including a full list of MNI coordinates and functional network labels.
Please cite: Seitzman BA, Gratton C, Marek S, Raut RV, Dosenbach NUF, Schlaggar BL, Petersen SE, Greene DJ. A set of functionally defined brain regions with improved representation of the subcortex and cerebellum.
Midnight Scan Club
The Midnight Scan Club (MSC) data set contains 10x high-fidelity individual connectomes (24-34 yo; 5F). For each participant 5 hr of RSFC data, 6 hr of task fMRI data, 4x T1, 4x T2, 4x MRA, 4x MRV and neuropsychological assessments including the NIH toolbox are available. All MRI data were collected after midnight to control for time of day effects. The data are freely available in raw format from openfmri.org and in processed format from neurovault.org.
Please cite: Gordon EM, Laumann TO, Gilmore AW, Newbold DJ, Greene DJ, Berg JJ, Ortega M, Hoyt Drazen C, Gratton C, Sun H, Hampton JM, Coalson RS, Nguyen A, McDermott KB, Shimony JS, Snyder AZ, Schlaggar BL, Petersen SE, Nelson SM, Dosenbach NUF. Precision functional mapping of individual human brains. Neuron. 2017;95(4):791-807.
Motion Feedback
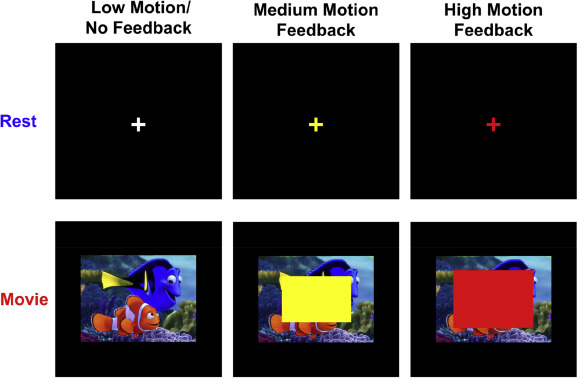
A major limitation to structural and functional MRI (fMRI) scans is their susceptibility to head motion artifacts. Even submillimeter movements can systematically distort functional connectivity, morphometric, and diffusion imaging results. In patient care, sedation is often used to minimize head motion, but it incurs increased costs and risks. In research settings, sedation is typically not an ethical option. Therefore, safe methods that reduce head motion are critical for improving MRI quality, especially in high movement individuals such as children and neuropsychiatric patients. We investigated the effects of (1) viewing movies and (2) receiving real-time visual feedback about head movement in 24 children (5-15 years old). Children completed fMRI scans during which they viewed a fixation cross (i.e., rest) or a cartoon movie clip, and during some of the scans they also received real-time visual feedback about head motion. Head motion was significantly reduced during movie watching compared to rest and when receiving feedback compared to receiving no feedback. However, these results depended largely on age, such that the effects were driven by the younger children. Children older than 10 years showed no significant benefit. We also found that viewing movies significantly altered the functional connectivity of fMRI data, suggesting that fMRI scans during movies cannot be equated to standard resting-state fMRI scans.
The implications of these results are twofold:
(1) given the reduction in head motion with behavioral interventions, these methods should be tried first for all clinical and structural MRIs in lieu of sedation.
(2) For fMRI research scans, these methods can reduce head motion in certain groups, but investigators must keep in mind the effects on functional MRI data.
Please cite: Greene DJ, Koller JM, Hampton JM, Wesewich V, Van AN, Nguyen AL, Hoyt C, McIntyre L, Earl EA, Klein RA, Shimony JS, Petersen SE, Schlaggar BL, Fair DA, Dosenbach NUF. Behavioral interventions for reducing head motion during MRI scans in children. Neuroimage. 2018; Jan 11.
Motion Feedback Videos
These custom video clips were used to present to child participants during resting-state scans in our study investigating movie clips and feedback on head motion. The clips consist of cartoon blockbuster movies edited for our specific research purposes. Three movies were used to make a total of seven movie clips that were shown to participants in a randomized order. Two clips were taken from Big Hero 6 (Disney Movies), two were from Despicable Me (Illumination Entertainment, Universal Pictures), and three were from Finding Nemo (Disney Movies, Pixar). Clips were chosen on the basis of being engaging, but not overly exciting or upsetting. Clips are approximately 7 minutes each corresponding to the length of the scans. We found that viewing movies significantly altered the functional connectivity of fMRI data, suggesting that fMRI scans during movies cannot be equated to standard resting-state fMRI scans. However, movie watching can reduce head motion in certain groups and should be used in clinical and structural MRIs without issue. For fMRI scans, investigators must keep in mind the effects on the data.
Please cite: Greene DJ, Koller JM, Hampton JM, Wesewich V, Van AN, Nguyen AL, Hoyt C, McIntyre L, Earl EA, Klein RA, Shimony JS, Petersen SE, Schlaggar BL, Fair DA, Dosenbach NUF. Behavioral interventions for reducing head motion during MRI scans in children. Neuroimage. 2018; Jan 11.
software
Dosenbach and Greene Lab GitHub
A link to the lab GitHub. Various tools our lab uses/develops are here.
Midnight Scan Club Code
Code for analysis of the MSC dataset
This codebase includes T1 segmentation, surface-based registration, myelin map creation, BOLD preprocessing, BOLD fc-processing, cifti creation, infomap-based network detection, gradient-based parcellation, and data reliability testing.
This site was designed and coded by Andrew Van. View the source code here. Copyright (©) 2018 Andrew Van.